[Guest post written by John Leavitt, Ph.D., retired Senior Scientist at LPISM in Palo Alto CA from 1981 to 1988; living in Woodstock CT.]

In 1980, Klaus Weber at the Max-Planck Institute and I published the amino acid sequence of human beta- and gamma-cytoplasmic actins. In 1981, after we completed this work, Klaus asked me “What are you going to do next?” I told him that I was moving to the Linus Pauling Institute of Science and Medicine in Palo Alto, California, and that I was going to clone the human beta-actin gene. My reason was that I had discovered a mutation in beta-actin that was associated with a tumorigenic human fibrosarcoma cell line. I wanted to test the hypothesis that this mutation contributed to the tumorigenic potential of this fibrosarcoma.
In 1984, I published the cloning of multiple copies of both the normal (wildtype) human beta-actin gene and multiple copies of the mutant gene. These actins are the most abundant proteins of all replicating mammalian cells and most other cells, down to yeast. (My story of meeting Dr. Pauling, moving from the National Institutes of Health to the LPISM, and our work on the role of this actin mutation in tumorigenesis in our model system was recounted in an article posted at the Pauling Blog in 2014.) In 2013, Schoenenberger et al. at the Biozentrum in Basel, Switzerland, reproduced all of our findings in a different cell system, a rat fibroblast model system, and extended our findings (see our review of their work).
A year ago, in June 2015, Dugina et al published a paper that proposed that altering the ratio of these two actins regulated either suppression or promotion of cancerous cell growth (more work needs to be done). I was very surprised by this idea – even though our work at LPISM had suggested this, I hadn’t thought of putting our observations into the language of “tumor suppression” and “tumor promotion.” Perhaps this was because, in the 1980s, hundreds of so-called “oncogenes” (tumor promoters) and tumor suppressor genes were being cataloged, and our findings were suggesting that a so-called “housekeeping” gene could do the same.
Indeed, Dugina and colleagues even stated this, somewhat simplistically, at the beginning of their Discussion section if their paper:
Until recently non-muscle cytoplasmic β- and γ-actins were considered only to play structural roles in cellular architecture and motility. They (the two isoforms) were viewed as products of housekeeping genes and β-actin was commonly used as internal control in various biochemical experiments.
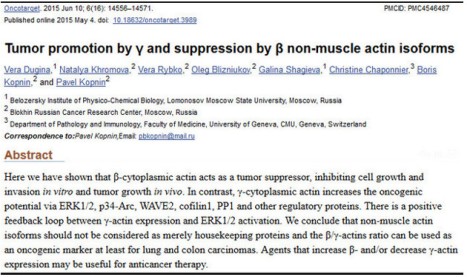
It didn’t go unnoticed by me that this paper failed to cite any of our papers, which had produced fundamental knowledge about human cytoplasmic actins. For example, instead of citing our 1980 paper on the amino acid sequences of human cytoplasmic beta- and gamma-actins, the Russian authors cited a paper on the sequences of bovine actins. Furthermore, these authors were apparently unaware of our discovery of actin mutations leading to tumorigenesis and several examples of null alleles of human beta-actin genes associated with tumors.
I communicated by email with the senior author of this paper, Pavel Kopnin at the Blokhin Russian Cancer Research Center in Moscow, not to complain about these omissions, but to tell him that I liked his hypothesis and to explain why. He thanked me and opined that he had had trouble persuading reviewers to publish the paper. I told him that our findings supported his hypothesis and would have made his argument stronger. He apologized for not citing our work and said that he had not reviewed the literature that far back, which amounted to twenty-eight years since our last paper from LPISM was published in 1987 (this made me feel old).
As early as March 1980, I had suggested in writing that altering the ratio of beta- and gamma-actins might contribute to the causation of cancer. This paper was published in the major journal, Journal of Biological Chemistry (see the figure below, last sentence of the abstract). If Dugina et al. were to consider filing a patent on this idea as an invention, our paper would have to, at least, be considered as invalidating prior art along with the rest of our work at LPISM up to 1987.

Both our work at LPISM and Schoenenberger’s work in Basel indicate that the mutation in one of two alleles of the beta-actin gene produces a stable, but defective, form of beta-actin. If Dugina’s hypothesis is correct, it is tempting to suggest that the function of the mutation site in beta-actin controls suppression of tumor formation. I recommended to Pavel Kopnin that his lab pursue this and it is my impression that his lab will continue to work on this hypothesis.
In our model system, we isolated a derivative cell line from the original mutated human fibrosarcoma cell line that exhibited even faster tumor formation (Leavitt et al, 1982). In this second cell line, the mutant beta-actin gene had acquired two additional mutations that made the mutant beta-actin labile with a fast turnover rate in the cell (Lin et al, 1985). As the result of this change, the ratio of stable beta- to gamma-actin changed from approximately 2:1 to approximately 1:1. Furthermore, we found that the two remaining stable forms of beta- and gamma-actin up-regulated in synthesis to maintain a constant normal amount of actin in the cell.
In addition, when we transferred additional mutant human beta-actin genes into immortalized but non-tumorigenic human fibrosarcoma cells, we found that both beta- and gamma-actin from the endogenous normal genes were down-regulated to maintain a constant stable amount of actin in the cell. Thus, we found and reported that beta- and gamma-actin levels in living cells auto-regulated the activities of their own endogenous genes to maintain a constant level of actin in the cell along with a constant ratio of these actins as well (Leavitt et al, 1987a; and Leavitt et al, 1987b). This finding was later confirmed by other laboratories.
These final observations lend support to the idea that maintaining a normal ratio of fully functional cytoplasmic beta- and gamma-actins may be required for the maintenance of the normal, non-neoplastic cellular phenotype. By contrast, mutations and deletions that alter the ratio of functional cytoplasmic beta-actin to gamma actin could lead to tumorigenesis. Hopefully, Pavel Kopnin and others who are aware of our work at LPISM will explore further the role of cytoplasmic actins in maintenance of the normal, non-neoplastic state.
Filed under: Colleagues of Pauling, Linus Pauling Institute | Tagged: beta-actin gene, cancer, John Leavitt | Leave a comment »